1. Introduction
Some info about GDI here.1.1. Goals and Scope of the Harmonized Minimal Data Model
The Harmonized Minimal Data Model (HMDM) has multiple purposes, some of these are:
-
Each dataset that is submitted to the catalogue should adhere (to a certain extend, still to be decided upon) to this model
-
Within the catalogue based on our model search options can be made available (filters/facets)
-
The HMDM can/needs to be implemented in the Beacons (and/or other discovery tools).
Next to the HMDM we provided a minimal metadata for submission schema.
Version 1 of metadata for submission model was released and can be seen here. This version was based on the version of HealthDCAT-AP from December 2024. Since the work on HealthDCAT-AP is still ongoing, there have been some changes since that time. This means our metadata for submission is not compliant to the current version of HealthDCAT-AP anymore. We will update the metadata for submission to version 1.1, which will be, according to HealthDCAT-AP, provided in 3 variants, based on the access level of the dataset: Open, Sensitive and Protected, which differ in cardinalities of the properties. The metadata for submission model will include all the current mandatory fields from those HealthDCAT-AP vairants, as well as a custom class created specifically for the submission of metadata in GDI, called HMD Submission. The name spaces for HMD Submission class and its properties were made available through FAIR Genomes repository. There are also some mandatory fields that were already included in the GDI MS8 deliverable, which will stay mandatory for backwards compatibility. This update will be very minimal, so only extending the current model with new mandatory fields.
1.2. Overview and Diagram
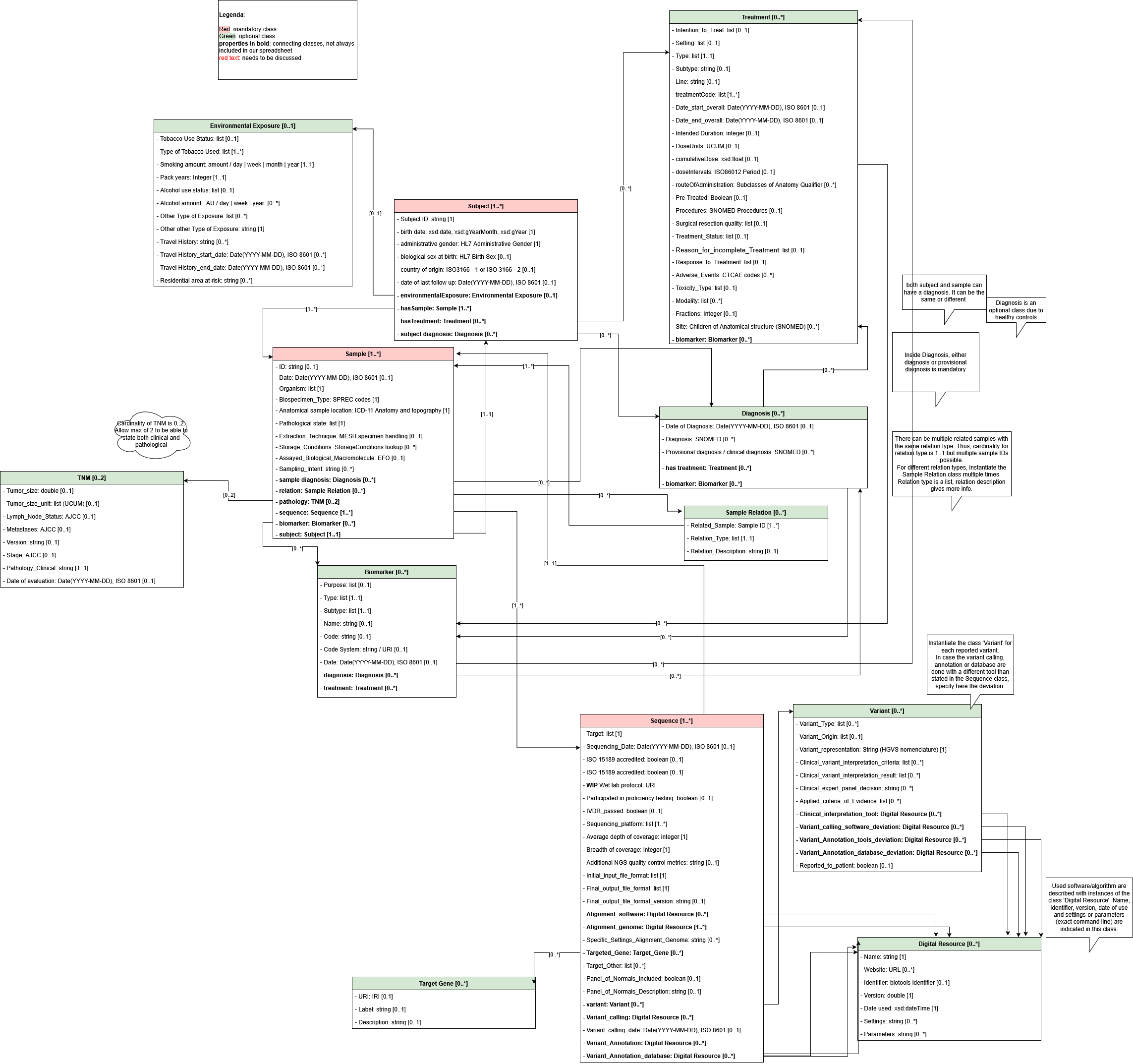
2. Main Classes
In the Harmonized Minimal Dataset, we defined 12 different classes.2.1. Subject
| Item | Definition | Value | Cardinality |
|---|---|---|---|
| Birth Date | The calendar date on which a person was born. | Complete date, without time, following the ISO 8601. If only year or year-month is available, use that. xsd:date or xsd:gYearMonth or xsd:gYear | 1..1 |
| Administrative Gender | The gender of a person used for administrative purposes. | HL7 Administrative Gender | 1..1 |
| biological sex at birth | The sex of a person at birth as molecular proven. | HL7 ValueSet: Birth Sex | 0..1 |
| Date_of_Last_Follow_up | Date of last follow-up, partial date with month and year. | Date (YYYY-MM-DD), ISO 8601 format | 0..1 |
| Country of Origin | A person’s descent or lineage, from a person or from a population. | 2- or 3-lettercode from ISO 3166-1 if only a country code is provided. If a country-subdivision then a value from the ISO 3166-2 | 0..1 |
| Subject ID | A sequence of characters used to identify, name, or characterize a trial or study subject. | String | 1..1 |
2.2. Diagnosis
| Item | Definition | Value | Cardinality |
|---|---|---|---|
| Date of Diagnosis | Date at which diagnosis was made. | Date (YYYY-MM-DD), ISO 8601 format | 0..1 |
| Diagnosis | The investigation, analysis and recognition of the presence and nature of disease, condition, or injury from expressed signs and symptoms; also, the scientific determination of any kind; the concise results of such an investigation. | Children of SNOMED-CT Disease (Disorder) | 0..n |
| Provisional diagnosis / clinical diagnosis | An initial diagnosis that is subject to change as new information becomes available. | Children of SNOMED-CT Disease (Disorder) | 0..n |
2.3. Sample
| Item | Definition | Value | Cardinality |
|---|---|---|---|
| Anatomical sample location | Anatomic site from which the sample was taken. | ICD-11 Anatomy and topography | 1..1 |
| Pathological state | The pathological condition of the sample. | Tissue Normal, Germline Normal, Primary Tumor, Tumor Metastasis, Recurrent Tumor, Organoid, Tumoroid | 1..1 |
| Date | Defines the date of sampling. | Date (YYYY-MM-DD), ISO 8601 format | 0..1 |
| ID | Unique identifier for a collected specimen assigned by data provider. | String | 0..1 |
| Organism | A living entity. | Children of NCIT Organism | 1..1 |
| Biospecimen_Type | The type of a material sample taken from a biological entity for testing, diagnostic, propagation, treatment or research purposes. This includes particular types of cellular molecules, cells, tissues, organs, body fluids, embryos, and body excretory substances. | Value from type of Sample table in SPREC Codes v3.0 :
ASC - Ascites fluid AMN - Amniotic fluid BAL - Bronchoalveolar lavage BLD - Blood (whole) BMA - Bone marrow aspirate BMK - Breast milk BUC - Buccal cells BUF - Unficolled buffy coat, viable CEL - Ficoll mononuclear cells, viable CEN - Fresh cells from non blood specimen type CLN - Cells from nonblood specimen type (e.g., disrupted tissue), viable CRD - Cord blood CSF - Cerebrospinal fluid NAS - Nasal washing PEL - Ficoll mononuclear cells, nonviable PEN - Cells from nonblood specimen type (e.g., disrupted tissue), nonviable PFL - Pleural fluid PL1 - Plasma, single spun PL2 - Plasma, double spun SAL - Saliva SEM - Semen SER - Serum SPT - Sputum STL - Stool SYN - Synovial fluid TER - Tears U24 - 24-h urine URN - Urine ZZZ Other | 1..1 |
| Extraction_Technique | The technique of extraction of the sample. | Values within MESH Specimen Handling | 0..1 |
| Storage_Conditions | Storage conditions of the sample. | FAIR Genomes Storage Conditions | 0..n |
| Assayed_Biological_Macromolecule | Macromolecule derived from the sample. | Children of EFO biological macromolecule | 0..1 |
| Sampling_Intent | Describes the purpose for taking the sample. | String | 0..n |
2.3.1. TNM
| Item | Definition | Value | Cardinality |
|---|---|---|---|
| Tumor_size | Indicates the size of the primary tumor. | Double | 0..1 |
| Tumor_size_unit | Indicates the unit in which tumor size is stated. | UCUM | 0..1 |
| Lymph_Node_Status | Indicates lymph node involvement according to the international TNM classification for solid tumors. | Children of SCTID: 1279504008 | American Joint Committee on Cancer ycN category allowable value (qualifier value) | 0..1 |
| Metastases | Indicates presence or absence of metastasis according to the international TNM classification for solid tumors. | Children of SCTID: 1222587001 | American Joint Committee on Cancer pathological M category allowable value (qualifier value), or Children of SCTID: 1222591006 | American Joint Committee on Cancer clinical M category allowable value (qualifier value), or HL7 NULL flavor in case not determined. | 0..1 |
| Version | Version of the TNM classification. | String | 0..1 |
| Stage | The extent of a cancer in the body. Staging is usually based on the size of the tumor, whether lymph nodes contain cancer, and whether the cancer has spread from the original site to other parts of the body. | Children of
SCTID: 1222592004 | American Joint Committee on Cancer clinical stage group allowable value (qualifier value), or SCTID: 1222593009 | American Joint Committee on Cancer pathological stage group allowable value (qualifier value), or SCTID: 1222594003 | American Joint Committee on Cancer yp stage group allowable value (qualifier value), or UICC values (to be added to SNOMED in 2025) | 0..1 |
| Pathology_Clinical | Indication whether TNM classification is based on clinical or pathological evaluation. If pathological is available, this always goes before clinical. | Clinical or pathological | 1..1 |
| Date of evaluation | Date of the clinical/pathological evaluation. | Date (YYYY-MM-DD), ISO 8601 format | 0..1 |
2.3.2. Sample Relation
| Item | Definition | Value | Cardinality |
|---|---|---|---|
| Related_Sample | Points to a related sample. | Sample ID | 1..n |
| Relation_Type | Describes the relationship type between the two connected samples. | List to be defined/extended:
New sample Derived Aliquot Control (experiment) Disease versus Control Matched Longitudinal Paired Familial Spatial | 1..1 |
| Relation_Description | Free text describing the Relation Type of two samples. | String | 0..1 |
2.4. Sequence
| Item | Definition | Value | Cardinality |
|---|---|---|---|
| Target | Identification of the sequenced target. | Whole Genome Sequencing (WGS) Whole Exome Sequencing (WES) Multi-gene panel sequencing array OTH | 1..1 |
| Sequencing_Date | Defines the date of sequencing. | Date (YYYY-MM-DD), ISO 8601 format | 0..1 |
| ISO 15189 accredited | Indication whether the laboratory is accredited according to ISO 15189 (clinical). | Boolean | 0..1 |
| ISO 17025 accredited | Indication whether the laboratory is accredited according to ISO 17025 (research). | Boolean | 0..1 |
| WIP Wet lab protocol | String or https://www.protocols.io URL | ||
| Participated in proficiency testing | Indication whether the laboratory had participated in any proficiency testing, such as interlaboratory comparison. | Boolean | 0..1 |
| IVDR_passed | Indicate whether the methodology (including chemistry and sequencing standards) used for sequencing follows the In vitro diagnostic medical devices (IVDR) regulation passed by the EU in April 2017. | Boolean | 0..1 |
| Sequencing Platform | The used sequencing platform (i.e. brand, name of a company that produces sequencer equipment). | FAIR Genomes or EFO list | 1..n |
| Average depth of coverage | Mean coverage for whole genome sequencing, or mean target coverage for whole exome and targeted sequencing (eg 60x, average number of times each target base has been ‘read’ by sequencer). | Integer | 1..1 |
| Breadth of coverage | Breadth of coverage (or evenness) is the proportion or percentage of a reads that has been sequenced at a the provided average depth of coverage. (E.g. if for Average depth of coverage, the value is 60, and the Evenness is 50% at 60x, the value here will be 50). | Integer | 0..1 |
| Additional NGS quality control metrics | Statement of any additional NGS quality control metrics. | String | 0..1 |
| Initial_input_file_format | Identification of the genomic file format of the initial input file (eg. fastq, bam, cram). | EDAM’s file types and formats | 1..1 |
| Final_output_file_format | Identification of the genomic file format of the final output file (eg. vcf, gvcf). | EDAM’s file types and formats | 1..1 |
| Final_output_file_format_version | Identification of the version of genomic file format of the final output file (eg. vcf, gvcf). | String | 0..1 |
| Alignment_software | Identification of the software used for alignment. | --> Digital Resource class | 0..n |
| Alignment_Genome | The specific build of the human genome used as reference for sequence alignment and variant calling. | --> Digital Resource class | 1..n |
| Specific_Settings_Alignment_Genome | Any specific settings regarding alternative contigs or decoys. | String | 0..n |
| Target Gene | In case of targeted sequencing, specify which gene is being targeted. This item points to another class: Target_Gene. | --> Target Gene class | 0..n |
| Target Other | Any other targeted genomic region. | Children of SIO region NULL flavors | 0..n |
| Panel_of_Normals_Included | Indicate whether a panel of normals is included during variant calling. | Boolean | 0..1 |
| Panel_of_Normals_Description | Free text description of panel of normals, if applicable. | String | 0..1 |
| Variant | A detected and reported variant. | --> Variant class | 0..n |
| Variant_calling | Identification of the software used for variant calling. | --> Digital Resource class | 0..n |
| Variant_calling_date | Defines the date of variant calling. | Date (YYYY-MM-DD), ISO 8601 format | 0..1 |
| Variant_Annotation | Identification of the software used for variant annotation. | --> Digital Resource class | 0..n |
| Variant_Annotation_database | Database and version used for variant annotation. | --> Digital Resource class | 0..n |
2.4.1. Digital Resource
| Item | Definition | Value | Cardinality |
|---|---|---|---|
| Name | The name of the tool/software/database used. | String | 1..1 |
| Website | Link to the website or repository (like GitHub) of the tool/software/database. | URL | 0..n |
| Identifier | bio.tools identifier for the digital resource. | bio.tools identifier | 0..1 |
| Version | The version of the tool/software/database used. | double | 1..1 |
| Date used | The date when the tool/software/database was last used. | xsd:dateTime | 1..1 |
| Settings | Free text account of the settings used in the tool/software/database. | String | 0..n |
| Parameters | Description of parameters used with the specified software. Copy the complete command line (all lines executed) used. | String | 0..n |
2.4.2. Variant
| Item | Definition | Value | Cardinality |
|---|---|---|---|
| Variant_Type | The category or type of variation or abnormality present in an amino acid or nucleic acid sequence. | SNVs, indels, SVs, CNVs, gene fusions, ... (to be extended). | 0..n |
| Variant_Origin | A quality inhering in a variant by virtue of its origin. | somatic, germline, maternal, paternal, pedigree specific, population specific, de novo. | 0..1 |
| Variant_representation | The representation of the variant using HGVS nomenclature. | String following HGVS nomenclature | 1..1 |
| Clinical_Variant_Interpretation_criteria | Internationally (e.g. ACMG, ESMO-ESCAT) criteria met for variant interpretation | List of versions of ACMG, ESMO-ESCAT, others?? | 0..n |
| Clinical_Variant_Interpretation_result | Indicates result of clinical variant interpretation. | benign, likely benign, VUS, likely pathogenic, pathogenic. | 0..n |
| Clinical_expert_panel_decision | Decision by clinical expert panel concerning the variant interpretation | String | 0..n |
| Applied_Criteria_of_Evidence | A category which fits with categories provided by Expert panels or tools accepted in Clincial Practice. If such recommendations are not available the weighted categories provided by freely available tools would be acceptable. | As listed in tables 3 and 4 in Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology | 0..n |
| Clinical_Interpretation_Tool | Identification of the tool used for clinical interpretation | --> Digital Resource class | 0..n |
| Variant_calling_software_deviation | Identification of the software used for variant calling, if different from software stated in Sequence class. | --> Digital Resource class | 0..n |
| Variant_Annotation_tools_deviation | Identification of the software used for variant annotation, if different from software stated in Sequence class. | --> Digital Resource class | 0..n |
| Variant_Annotation_database_deviation | Database and version used for variant annotation, if different from software stated in Sequence class. | --> Digital Resource class | 0..n |
| Reported_to_patient | Indication if the variant has been reported back to the patient, if different from software stated in Sequence class. | Boolean | 0..1 |
2.4.3. Target Gene
| Item | Definition | Value | Cardinality |
|---|---|---|---|
| URI | URI identifying the targeted gene. | URI to either HGNC NCBI gene, OMIM, HPO or HGVS for variants. | 0..1 |
| Label | Label of the target gene, if no URI can be provided. | String | 0..1 |
| Description | Description of target gene. | String | 0..1 |
2.5. Treatment
| Item | Definition | Value | Cardinality |
|---|---|---|---|
| Intention_to_Treat | Indicate the intended disease outcome for which the treatment is given, may be coded as SNOMED-CT. | SCTID: 373808002 | Curative - procedure intent
SCTID: 373847000 | Neoadjuvant intent SCTID: 373846009 | Adjuvant - intent SCTID: 363676003 | Palliative - procedure intent SCTID: 360271000 | Prophylaxis - procedure intent SCTID: 243114000 | Support (regime/therapy) SCTID: 129304002 | Excision - action SCTID: 261004008 | Diagnostic intent SCTID: 129428001 | Preventive - intent SCTID: 429892002 | Guidance intent SCTID: 360156006 | Screening - procedure intent SCTID: 447295008 | Forensic intent Other - HL7 NULL flavor (OTH, UNK, NI, NA) | 0..1 |
| Setting | Indicate the treatment setting, which describes the treatment’s purpose in relation to the primary treatment (Reference: NCIT C124308) | Adjuvant Advanced/Metastatic Neoadjuvant Not applicable | 0..1 |
| Type | Indicates the type of treatment regimen that the patient completed; coded values can be chosen, SNOMED may be chosen for "procedures". | SCTID: 23719005 | Bone marrow transplant
SCTID: 367336001 | Chemotherapy SCTID: 18629005 | Medication SCTID: 423827005 | Endoscopic therapy SCTID: 169413002 | Hormonal therapy SCTID: 76334006 | Immunotherapy SCTID: 257891001 | Photodynamic therapy SCTID: 1287742003 | Radiation therapy SCTID: 56305001 | Radionucleotide therapy SCTID: 1269349006 | Stem cell transplant SCTID: 387713003 | Surgery SCTID: 394613000 | Gene therapy SCTID: 310341009 or 373818007 | Watchful waiting SCTID: 424313000 | Active follow-up SCTID: 703423002 | Chemoradiotherapy HL7 NULL flavor (NI, NA, OTH) | Other | 1..1 |
| Subtype | Detailed specification of the treatment type. | String | 0..1 |
| Line | Indicate if the treatment was the primary treatment following the initial diagnosis or 2nd, 3rd, … | String | 0..1 |
| treatmentCode | A drug product that contains one or more active and/or inactive ingredients used by the patient intended to treat, prevent or alleviate the symptoms of disease. Any hormone therapies, gender-related or otherwise, should also be recorded here. | ATC codes, IDMP when available. In case of Cancer: HEMONC, RxNorm | 1..n |
| Date_start_overall | The date and time of (the start of) the treatment. | Date (YYYY-MM-DD), ISO 8601 format | 0..1 |
| Date_end_overall | The date and time of the end of the overall treatment. | Date (YYYY-MM-DD), ISO 8601 format | 0..1 |
| Intended Duration | The duration of treatment regimen, in days. | Integer | 0..1 |
| DoseUnits | Indicates the total dose given in units (e.g. of Gray (Gy) when radiation, Millimeters/24hours for medication). | Choicelist with most common DoseUnits, like Gray, Milligrams/24 hours: make use of UCUM. | 0..1 |
| cumulativeDose | The amount of any substance administered over a specific period of time. | Float | 0..1 |
| doseIntervals | The number of times a substance is administered within a specific time period. | ISO8601 Period | 0..1 |
| routeOfAdministration | Designation of the part of the body through which or into which, or the way in which, the medicinal product is intended to be introduced. In some cases a medicinal product can be intended for more than one route and/or method of administration. | Subclass of Anatomy qualifier | 0..n |
| Pre-Treated | Indicates if disease has been pre-treated or is in course of treatment. | Boolean | 0..1 |
| Procedures | Indicates the type of treatment regimen that the patient completed; coded values can be chosen, SNOMED may be chosen for "procedures". | Children of SNOMED Procedure | 0..1 |
| Surgical resection quality | Evaluation of the surgical resection quality based on coded values. | SCTID: 1222638005 |American Joint Committee on Cancer R0 (qualifier value)
SCTID: 1222639002 | American Joint Committee on Cancer R1 (qualifier value) SCTID: 1222640000 | American Joint Committee on Cancer R2 (qualifier value) SCTID: 1222641001 | American Joint Committee on Cancer RX (qualifier value) | 0..1 |
| Treatment_Status | Indicates the patient’s outcome of the prescribed treatment (coded values, e.g. treatment not completed | because of toxicity). | SCTID: 182992009 | Treatment completed (situation)
SCTID: 445528004 | Treatment changed (situation) SCTID: 405613005 | Planned procedure (situation) SCTID: 416406003 | Procedure discontinued (situation) | 0..1 |
| Reason_for_incomplete_Treatment | Reason for discontinuation of the treatment. | SCTID: 419099009 | Dead (finding)
SCTID: 1296859006 | Procedure declined (situation) SCTID: 713247000 | Procedure discontinued by patient (situation) SCTID: 713246009 | Procedure discontinued by healthcare professional (situation) SCTID: 266721009 | Absent response to treatment (situation) SCTID: 407563006 | Treatment not tolerated (situation) technical or organizational problems | to be added, or use OTH Other | HL7 null flavour OTH Not applicable | HL7 null flavour, NA No information | HL7 null flavour, NI | 0..1 |
| Response_to_Treatment | The patients' response to the applied treatment regimen. (Source: RECIST). | Complete Response Disease progression NED Partial Response Stable Disease | 0..1 |
| Adverse_Events | Reports any treatment related adverse events. (Codelist reference: NCI-CTCAE (v5.0)) | CTCAE Codes | 0..n |
| Toxicity_Type | If the treatment was terminated early due to acute toxicity, indicates the type of toxicity that caused early termination of treatment. | Children of Toxicity (NCIT) | 0..1 |
| Modality | Indicates the method of radiation treatment or modality. | Electron Heavy Ions Photon Proton | 0..n |
| Fractions | Indicates the total number of fractions delivered as part of radiation treatment. | Integer | 0..1 |
| Site | Indicates the body region where radiation therapy was administered. | Children concepts of Anatomical Structure (body structure) | 0..n |
2.6. Biomarker
| Item | Definition | Value | Cardinality |
|---|---|---|---|
| Purpose | Necessary information to indicate the objective of the biomarker used (coded value, e.g. diagnostic, prognostic,…). | Diagnosis Prognosis Prediction Monitoring | 0..1 |
| Type | What type is the biomarker classified as. | Molecular
Imaging Anthropometric Cellular Physiological | 1..1 |
| Subtype | Subtype of the biomarker as classified in the type. | If Type ==
- Molecular: Genetics/Genomics, Epigenetics/Epigenomics, Transcription/Transcriptomics, Metabolites/Metabolomics, Proteins/Proteomics, Microbiomics/Microbiology, Biochemistry (biochemical), Other molecular biomarker, N/P - Imaging: X-Rays, Ultrasound (echography, etc), CT Scan, PET/SPECT, Spectrometry, MRI, Scintigraphy (Gamma), Mammography, Other image biomarker, N/P - Anthropometric: BMI, Body perimeters (circumference), Other anthropometric biomarker, N/P - Cellular: Histology (tissue abnormalities), Cytology (cell types), Other cellular biomarker, N/P - Physiological: Blood Pressure, Ankle-brachial Index, ECG, EEG, Electromyography, Other physiological biomarker, N/P | 1..1 |
| Name | Biomarker name. | String | 0..1 |
| Code | Code of the biomarker. | String | 0..1 |
| Code System | Code System of the Biomarker Code. | String or URI | 0..1 |
| Date | The date of the biomarker being obtained. | Date (YYYY-MM-DD), ISO 8601 format | 0..1 |
2.7. Environmental Exposure
| Item | Definition | Value | Cardinality |
|---|---|---|---|
| Tobacco Use Status | The status of the patient’s tobacco use. | SCTID:449868002 | Smokes tobacco daily
SCTID:428041000124106 | Occasional tobacco smoker SCTID:43381005 | Passive smoker SCTID:8517006 | Ex-smoker SCTID:405746006 | Current non smoker but past smoking history unknown SCTID:266919005 | Never smoked tobacco NullFlavor OTH | 0..1 |
| Type Of Tobacco Used | Type of tobacco the patient uses. | Children of tobacco product combined with childred of vaporizer, or
HL7 NULL flavors | 1..n (conditional) |
| Smoking amount | The number of cigarettes, cigars or grams of rolling tobacco consumed per day, week, month or year. | amount/day | amount/week | amount/month | amount/year | 1..1 (conditional) |
| Pack years | The unit indicating the smoker’s total exposure to tobacco smoke. For cigarettes, this is calculated using the number of smoked packs of cigarettes per day (one pack = 20 cigarettes) times the number of years of smoking. For other forms of tobacco, this is usually converted to an equivalent cigarette consumption. Often, only the number of pack years is estimated. | Integer | 1..1 (conditional) |
| Alcohol use status | The status of the patient’s alcohol use. | SCTID:219006 | Current drinker
SCTID:105542008 | Non - drinker SCTID:82581004 | Ex-drinker SCTID:783261004 | Lifetime non-drinker of alcohol NullFlavor OTH | 0..1 |
| Alcohol amount | The extent of the patient’s alcohol use in units of alcohol per time period (day/week/year). | AU per (day/week/year). 1 A.U.=
125 ml of wine, 330 ml of beer, 80 ml of drink, 40 ml liquor | 0..n |
| Other Type of Exposure | Indicates other exposures other than tabacco/smoking and alcohol use, for example asbestos. This exposure can include direct physical contact, inhalation, ingestion, or residing in close proximity to the source. | - Inadequate water (quantity and quality), sanitation and solid waste disposal, improper hygein (handwashing)
- Improper water resource managment, uncluding poor drainage, - Crowded housing and poor ventialtion of smoke - Exposures to vehicular and industrial air pollution - Population movement and encroachment and construction, which affect feeding and breeding grounds of vectors, such as mosquitoes, - Exposure to naturally ocurring toxic substances - Natural resources degradation (for example, landslides, poor drainage, erosion) - Climate change, partly from combustion of fossil fuel and release of greenhouse gases in transportation, industry, and poor energy conservation in housing, fuel, commerce, and industry - Ozone depletion from industrial and commercial actvitiy Biological hazard (Bacteria, Fungi, Parasitic worms, Protozoa, Viruses, Prions) Chemical hazards (Air pollutant, Heavy metal (Arsenice, Mercury, Lead), Pesticides, Other ( Formaldehyde, Asbestos, PFAS, PCBs, BPA, Phthalates, Radon, DDT) Physical environmental hazard (Radiation, Natural disaster (narrow matches: exposure to earthquake, exposure to flooding, exposure to tsunami etc.), Extreme weather, Human activities (eg. traffic accident)) HL7 NULL flavors | 0..n |
| Other_Other Type of Exposure | Provide the option to if "Other" option is choosen from List of other exposures, type a free text field. | String | 1..1 (conditional) |
| Travel History | Description of any travel within 4 weeks before the diagnosis. | String | 0..n |
| Travel History_start_date | Start date of relevant travel history. | Date (YYYY-MM-DD), ISO 8601 format | 0..n |
| Travel History_end_date | End date of relevant travel history. | Date (YYYY-MM-DD), ISO 8601 format | 0..n |
| Residential area at risk | Description of the area of known environmental exposure conditions where the subject/patient resides. | String | 0..n |
3. Matches
Different countries use different standards or vocabularies for their data. For example, in the harmonized minimal data model we mostly use terms from SNOMED CT and NCIT, but some countries only use ICD-10, and it’s difficult for them to convert everything to SNOMED. To make things easier, we decided to focus on matches between terms from different systems. This way, a dataset using ICD-10 can still be included in the GDI Catalogue, as long as we can clearly show how its terms connect to SNOMED or other terms.
For example:
-
One dataset might use a SNOMED term to describe a disease.
-
Another dataset might describe the same disease using an ICD-10 term.
-
Without a match between them, our system would think they are two different things.
-
If we add an exact match, the system will understand that they are the same, just described with different vocabularies.
This will make data both human and machine readable.
3.1. How To Use Matches
We started by adding different types of matches to all items and values in the model. The types of matches (match attribute), their definitions and examples can be found in the table below.
| Match Attribute | Definition | Example |
|---|---|---|
| skos:exactMatch | Two concepts have the same meaning and can be used interchangeably in all contexts. This link is transitive, meaning if A is an exact match to B, and B is an exact match to C, then A is also an exact match to C. | SNOMED Date of birth has an exact match to EFO Date of birth. Both refer to the same concept with no variation in meaning. |
| skos:closeMatch | Two concepts are sufficiently similar that they can be used interchangeably in many applications and schemes, but they are not strictly identical. This link is not meant to be transitive. | SNOMED Patient has a close match to NCIT Study Participant. A patient is an individual receiving medical care, while a study participant is someone enrolled in a research study. In some cases, these terms can be used interchangeably (e.g., in clinical trials involving patients), but not all study participants are patients—some might be healthy controls. |
| skos:relatedMatch | Two concepts are associated but not similar enough to be considered exact or close matches. | The concept HGNC BRCA1 and NCIT Breast Carcinoma have a skos:relatedMatch relationship because they are strongly associated but not identical. BRCA1 is a gene mutation, while breast cancer is a disease that may develop as a result of certain BRCA1 mutations. However, not all BRCA1 mutations lead to breast cancer, and not all cases of breast cancer are caused by BRCA1 mutations, making them related but not broader/narrower concepts. |
| skos:broadMatch | The concept represented by the external entity is broader than the item. This is a reverse property of skos:narrowMatch. | The concept NCIT Breast Carcinoma has a skos:broadMatch relationship to SNOMED Cancer. This means that Cancer is the broader concept because it represents a general category that includes multiple types of cancer, one of which is Breast Cancer. |
| skos:narrowMatch | The concept represented by the external entity is narrower than the item. This is a reverse property of skos:broadMatch. | The concept SNOMED Cancer has a skos:narrowMatch relationship to NCIT Breast Carcinoma. This means that Breast Cancer is a specific subtype of Cancer. |
Next to the matches, we also record what is the source of the match. If the mapping has already been done, for example SNOMED to ICD-10 in the SNOMED browser, or in the T-Rex browser, we include this information with the match in the spreadsheet.
3.2. Using SSSOM
From now on, we decided to follow the SSSOM (Simple Standard for Sharing Ontological Mappings) method. This is a standard format for describing matches between terms. We will:-
Add all required SSSOM elements.
-
Include recommended and optional elements if they are useful.
-
Make sure our matches follow the SSSOM rules.
Using this model and the SSSOM standard will help us:
-
Make it easier for teams using different vocabularies to share their data.
-
Let people and software search and analyze the data more effectively.
-
Support more countries and teams to join the GDI Catalogue without needing to change everything.
